EGAP v3 is Here: Powering Genome Assembly for Fungi and Beyond
- Ian Bollinger
- May 9, 2025
- 3 min read
TL;DR
EGAP v3 is a containerized, reproducible genome assembly pipeline built with Python-driven modules for metadata parsing and QC, hybrid assembly tools, and step-wise polishing and curation, orchestrated via Nextflow DSL2 for portability and scalability. It supports Docker and Singularity environments for consistent execution across platforms, is available through one-line installation on Bioconda, and was proven in assembling Psilocybe genomes in 2024 for a Microbiology Resource Announcements publication featuring new-to-science organisms.
Hello, genomics enthusiasts! I’m thrilled to share some exciting news: the newest release of Entheome Genome Assembly Pipeline (EGAP)! As a passionate bioinformatician, I’ve poured my heart into making EGAP a go-to tool for hybrid genome assembly, and this latest version is a game-changer for fungal genomes—and beyond. Whether you’re working with Oxford Nanopore (ONT), Illumina, or PacBio data, EGAP v3 delivers high-quality assemblies with ease and precision. Let’s dive into what makes this release so special!
What is EGAP?
EGAP is a custom-built bioinformatics pipeline for hybrid genome assembly. It handles everything from preprocessing raw sequencing data to assembling, polishing, and evaluating genomes. Supporting inputs like Illumina-only, Illumina + ONT, or PacBio-only, EGAP is optimized for fungal genomes but flexible enough for bacteria, plants, and other eukaryotes. It uses gold-standard tools like MaSuRCA, Flye, SPAdes, BUSCO, and QUAST to produce assemblies ranked as AMAZING, GREAT, OK, or POOR based on metrics like BUSCO Completeness and N50.
With EGAP v3, I’ve taken this pipeline to new heights, making it more powerful, user-friendly, and ready for your next big project.
What’s New in EGAP v3?
This release is packed with upgrades to streamline your workflow and boost assembly quality. Here’s what I’ve added:
1. Smarter Preprocessing
Better Read Handling: Merge FASTQ files and filter low-quality reads with Filtlong and Ratatosk (ONT) or Trimmomatic (Illumina).
Deduplication and Metrics: Use Clumpify to remove duplicates and get detailed read stats via FastQC, NanoPlot, and BBMap.
2. Enhanced Assembly and Selection
Multiple Assemblers: Choose MaSuRCA, Flye, SPAdes, or hifiasm, with EGAP picking the best assembly automatically.
Robust Evaluation: Dual-lineage BUSCO/Compleasm analysis and QUAST metrics (N50, contig count) ensure top-notch results.
3. Polishing and Curation
Refined Assemblies: Polish with Racon (long reads) and Pilon (Illumina), plus purge_dups to clean up haplotigs.
Scaffolding and Gap Closing: Use RagTag for reference-guided scaffolding and TGS-GapCloser or Abyss-Sealer to close gaps.
4. Easier Installation
Install via Conda (conda install -c bioconda egap), Docker, Singularity, or my handy EGAP_setup.sh script.
Containerized options make setup reproducible and hassle-free.
5. User-Friendly Features
Simple CSV Input: Just provide a CSV with your sample details (SRA, FASTQ paths, BUSCO lineages).
Customizable Runs: Adjust CPU threads and RAM to fit your system.
Clear Outputs: Get BUSCO plots, QUAST metrics, and intuitive assembly classifications.
Why EGAP v3 for Fungi and Beyond?
EGAP v3 is designed with fungal genomics in mind, but its flexibility makes it a powerhouse for any organism. Here’s why I think you’ll love it:
Hybrid Assembly Mastery: Combine short and long reads for superior results.
Fungal Focus: Optimized for fungi with customizable BUSCO lineages (e.g., basidiomycota, agaricales).
Ease of Use: From setup to results, EGAP is built to save you time.
Open-Source: Licensed under BSD 3-Clause, it’s free to use and improve.
Real Results
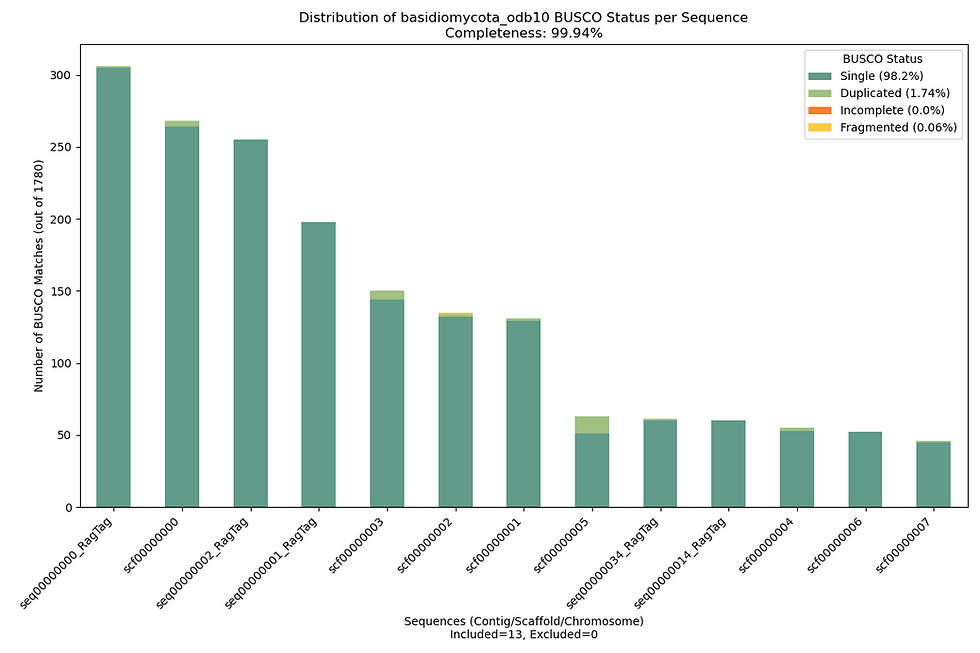
I’ve tested EGAP v3 on fungal species like Psilocybe cubensis and Panaeolus papilionaceus, achieving BUSCO completeness >98.5% in many cases. The included EGAP_test.csv lets you try it yourself—expect stunning BUSCO plots and detailed QUAST reports. Check out the repo for more examples of what v3 can do!
Join the Journey
EGAP is my labor of love, and I’d love for you to be part of its journey. Here’s how you can get involved:
Try It Out: Download EGAP v3 from GitHub and share your results.
Contribute: Have ideas for improvements? Submit a pull request or issue on GitHub.
Cite EGAP: Using it in your research? For now please reference EGAP and the awesome work by Bollinger et al. (2024) and Muñoz-Barrera et al. (2024); we are working getting its own publication going!
What’s Next?
I’m already dreaming up EGAP v4, with features like comprehensive quality reports and support for more genomes. Got suggestions? Drop them in the GitHub issues or let me know in the comments below!







Comments